[click on image to start the interactive model, NOTE: CPU intensive]
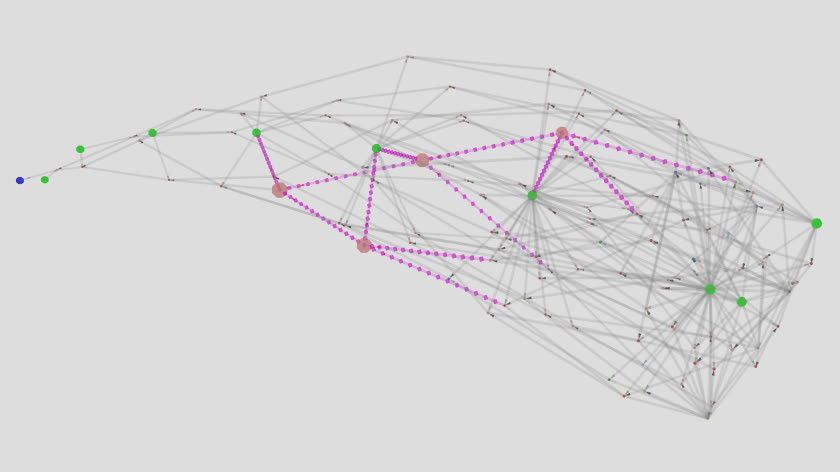 [A 5-node 9-variable (5n9) structure (magenta) spanning an expression.
[A 5-node 9-variable (5n9) structure (magenta) spanning an expression.
untangle-dataset
RockingShip proudly presents the third version of the dataset describing practical fractal space.
This is the first time the 4n9-mixed dataset is complete. Jay!
That means the detector/creator are fully seamless.
TL;DR
Grab compressed json file, unpack and build.
For latest 4n9-full:
wget https://rockingship.github.io/untangle-dataset/untangle-4n9-full-20211012.json.xz
xz -d untangle-4n9-full-20211012.json.xz
genimport untangle-4n9-full-20211012.db untangle-4n9-full-20211012.json
ln -s untangle-4n9-full-20211012.db untangle.db
For latest 4n9-mixed:
wget https://rockingship.github.io/untangle-dataset/untangle-4n9-mixed-20210926.json.xz
xz -d untangle-4n9-mixed-20210926.json.xz
genimport untangle-4n9-mixed-20210926.db untangle-4n9-mixed-20210926.json
ln -s untangle-4n9-mixed-20210926.db untangle.db
Areas
“Signatures” are to identify the function of structures.
“Members” are used to create fractal structures.
“Patterns” (not yet included) are used to detect fractal structures.
Members are always “safe”, meaning they are fully interlocking.
When overlapping multiple members, the overlap is also a safe members.
Using safe members to construct or extend fractal trees should (internally) never trigger normalisation.
There are 3 dataset areas:
-
Full, anything goes
Adding a
QTF/QnTF` nodes to trees will rewrite them to make the additions safe. -
Mixed, for creating
QnTF-only trees usingQTF/QnTF` (mixed) nodesSignatures are present for all possible top-level
QTF/QnTF(mixed) nodes. Members to create nodes to extend the tree areQnTF` only. -
Pure, for creating
QnTF-only trees usingQnTF-only nodesSignatures and members all assume
QnTF-only nodes. An additional advantage is that signatures can span larger structures.
Manifest
Available are json import to quickly construct the database. Also available are tarballs containing the build log and intermediate files.
Files hosted on this page (latest versions first):
-
Version 20210926 using untangle 2.11.0
This version was built with
--cascade --rewrite.-
untangle-4n9-full-20211012.json.xz - Import data
SIZE=1513232, MD5=f113a5c80ee9e17d611dc73016118cb4 -
untangle-4n9-full-20211012.tar.xz - Data files
SIZE=1292264, MD5=64c3c755bc9f69b2d984a2b21402527c
-
-
Version 20210926 using untangle 2.10.1
This version was built with cascading dyadics
--cascade.
Cascading is left-hand-side only (“ab^c^d^e^”).-
untangle-4n9-full-20210926.json.xz - Import data
Corrupted, please use the 20211012 version -
untangle-4n9-full-20210926.tar.xz - Data files
Corrupted, please use the 20211012 version -
untangle-4n9-mixed-20210926.json.xz - Import data
SIZE=683988, MD5=1d2c7671171560a9316c93284b082ff4 -
untangle-4n9-mixed-20210926.tar.xz - Data files
SIZE=558844, MD5=abaa3225f2344ebb08c869c18b981663
-
-
Version 20210810 using untangle 2.9.1
This version was built with cascading dyadics
--cascade-
untangle-4n9-full-20210810.json.xz - Import data
SIZE=1589212, MD5=13c364981bb76def331c8add85dc435f -
untangle-4n9-full-20210810.tar.xz - Data files
SIZE=1338416, MD5=de9c0132c082d2fe9f1b7cdb596c32e3 -
untangle-4n9-mixed-20210810.json.xz - Import data
SIZE=720628, MD5=90a96f1c00e78ffd07c5ff28773ba5b9 -
untangle-4n9-mixed-20210810.tar.xz - Data files
SIZE=1857848, MD5=45f4ee784018806a6baed8cef39fd4df
-
-
Version 20210808 using untangle 2.9.0
-
untangle-4n9-full-20210807.json.xz - Import data
SIZE=1523952, MD5=6f8d6705f48a8035dceb5a878018932e -
untangle-4n9-full-20210807.tar.xz - Data files
SIZE=1305940, MD5=6821fc28b4ee5d2efcf75f3defeb46b5 -
untangle-4n9-mixed-20210807.json.xz - Import data
SIZE=739708, MD5=d9bbf7993ea64955b1aaaf6a7cf7fb83 -
untangle-4n9-mixed-20210807.tar.xz - Data files
SIZE=1386808, MD5=f6f3889dc01ec44826d4867127188086
-
-
Version 20210723 using untangle 2.8.0
!! This version has a critical error in the member head/tail matching and should not be used !!
-
untangle-mixed.20210723.json.xz - Import data
SIZE=3909328, MD5=73fa36b5f2dcc2d7cf80ab0a94ab5d5a -
untangle-mixed.20210723.tar.xz - Data-list files
SIZE=3034856, MD5=4c9f0974d902dada332484cbe3fb3776 -
untangle-pure.20210723.json.xz - Import data
SIZE=1125616, MD5=e5b40b8dec4c866a1bcb63f4aa7022d0 -
untangle-pure.20210723.tar.xz - Data-list files
SIZE=857404, MD5=b8b8784c7b97272ccf44f263ab980b23
Other files/versions can be found on the github project page https://github.com/RockingShip/untangle-dataset
-
Table of contents
- Areas
- Manifest
- Table of contents
- Dataset
5n9-mixed - Dataset
6n9-pure - Building and Installation
untangle-mixed.dbfrom import fileuntangle-mixed.dbfrom list files (15 minutes)untangle-mixed.dbfrom scratch (2-3 hours)untangle-pure.dbfrom scratch (40 hours)- Requirements
- Versioning
- License
Dataset 5n9-mixed
Generating the dataset from scratch takes about 80 single-core hours to construct.
The essence is exported as a 100Mbyte JSON, compressed to 9M, which when imported and indexed will become 20G.
The dataset contains:
-
Transforms/skins (362880)
All 9! forward and reverse permutations of the 9 variables.
Specifically ordered to form a directional vector. -
Initial evaluator state, copy-on-write (1GByte)
For evaluating trees with 9-variables, using 9! 512-bit vectors
Generating the initial state takes about 15 CPU seconds.
Using copy-on-write reduces that to near-zero. -
Signatures (791646)
Normalised uniqueness of all
4n9structures. -
Swaps (151)
Variable/endpoint position reordering based on signature function.
Common situations are commutative properties of symmetrical operators. -
Imprints (181680347)
Associative tree lookup that determines its function (signature) and variable permutation (transform).
The index/data is 12Gbyte large. -
Members (6258677)
Instances used to detect and construct
5n9structures. All members are fully interlocking (safe) and independent (diversity).
If a member is broken into smaller fragments, each piece would also be “safe”.
Each member is used complete or partly as a component by other members.
Dataset 6n9-pure
Additionally available is the experimental dataset consisting exclusively of QnTF operators.
This database is 8GByte large.
The dataset is based on 4n9 pure signatures.
These are signatures where all nodes are exclusively QnTF except the top-level node that may be mixed QnTF or QTF.
All the nodes of member structures are exclusively QnTF.
Just like the main dataset, members of 5n9-pure space are used to fill blank spots of 4n9-pure space.
Only it turns out that 3 of the 193170 signatures have missing members, requiring to search for them in 6n9-pure space.
The challenge is that scanning 5n9-pure space takes 9 hours and 6n9-pure space is 450 times larger (4.1e11 structures).
Speed improvement is achieved by narrowing the search to only the missing signatures.
In addition, the lookup index is set to full-associative mode which is 720 times faster.
A full associative index is also 720 larger which makes it only feasible for tiny signature collections.
The dataset size is significantly different:
- Pure: 791646 signatures and 3651374 members
- Mixed: 193170 signatures and 381582 members
Building and Installation
There are three ways to build the database:
-
Importing (recommended)
Very fast and import is checksum protected.
-
Loading the data and re-applying validation takes about 15 minutes.
The advantage is that data can be tweaked, mixed or manually constructed. -
This can take days unless there is a High-Performance-Computing grid lying around.
Untanglewas build with the Sun-Grid-Engine in mind.
Build system
Untangle is designed to run and build using 32bit systems fitted with 32G memory.
Untangle is also heavily memory I/O bound and will likely kill CPU cache.
Effort has been done to access the evaluator and imprints in such a way to keep as much in cache as possible.
The database has been designed for grid environment where the database is hosted on a NFS share and loaded into the file system cache.
It is accessed using mmap() in copy-on-write mode and has a number of fallback options.
The database is explicitly accessed to not use swap-space or core-dump.
The current suggestion is a 64G/8-core system:
- 4G for linux
- 8G for applications
- 20G filesystem cache
- 32G (8x4G) for the workers
The idea is that the database is loaded, located and accessed in the file-system cache during the full run of a program.
If things go slow, and kswapd has been spotted, then most likely too many applications are running.
The database generator programs typically open the input database using mmap(),
copy the sections being modified or created to application memory,
then at the very last moment open the output database with posix_fadvise(POSIX_FADV_DONTNEED).
Saving a 18G database file can take between 10 seconds to several minutes, even on SSD.
untangle-mixed.db from import file
genimport untangle-mixed.db untangle-mixed.json
untangle-mixed.db from list files (15 minutes)
Export lists contain the values present in the database sections.
They are re-validated when loaded and are basically are a fast-forward without false-positives.
The advantage is that data can be examined, mixed and even modified.
Grab and unpack one of the data-list archives.
The following paragraph contain the commands to create and load them into the database.
Section transform
Create initial database containing pre-calculated mandatory sections.
[xyzzy@host v2.8.0]$ ./gentransform transform.db
Load signature
Load the list with signatures into the database.
Node size = 4.
[xyzzy@host v2.8.0]$ ./gensignature transform.db 4 signature.db --load=signature.lst --no-generate
Build swap
The variabl/endpoint swap information needs to be calculated.
[xyzzy@host v2.8.0]$ ./genswap signature.db swap.db
Load member
Load the list with members into the database.
Depreciated members have already been removed from the list.
Node size = 5.
[xyzzy@host v2.8.0]$ ./genmember swap.db 5 untangle-mixed.db --load=member.lst --no-generate
Creating data-lists
[xyzzy@host v2.8.0]$ ./gensignature untangle-mixed.db 4 --no-generate --text=3 > signature.lst
[xyzzy@host v2.8.0]$ ./genmember untangle-mixed.db 5 --no-generate --text=3 > member.lst
untangle-mixed.db from scratch (2-3 hours)
The following paragraphs contain the commands and screen logs to create the database section.
Building from scratch is a resource demanding experience.
Untangle developer environment has 32x 4G SGE grid nodes at its disposal.
On occasions, it has been extended to 128 nodes on Amazon S3.
Note that in case of storage shortage, older intermediate databases can be deleted.
Section transform (1 minute)
Create initial database containing pre-calculated mandatory sections
[xyzzy@host v2.8.0]$ ./gentransform transform.db
[00:00:00] Allocated 0.854G memory
[00:00:00] Generated 362880 transforms
[00:00:12] Writing transform.db
[00:00:12] Written transform.db, 854025184 bytes
Section signature (10 minutes)
Multi-pass creation of signatures, saving checkpoint data.
[xyzzy@host v2.8.0]$ ./gensignature transform.db 1 signature-1n9.db --text=1 >signature-1n9.ckp
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] Generating candidates for 0n9
[00:00:01] numSlot=9 pure=0 numNode=0 interleave=504 numCandidate=0 numSignature=3(33%) numImprint=6(5%) | skipDuplicate=0
[00:00:01] Generating candidates for 1n9
[00:00:01] numSlot=9 pure=0 numNode=1 interleave=504 numCandidate=6 numSignature=9(100%) numImprint=108(96%) | skipDuplicate=0
[00:00:01] Sorting signatures
[00:00:01] Rebuilding imprints
[00:00:01] Imprints built. numImprint=108(96%) | hash=1.065
[00:00:18] Writing signature-1n9.db
[00:00:18] Written signature-1n9.db, 854036704 bytes
[xyzzy@host v2.8.0]$ ./gensignature signature-1n9.db 2 signature-2n9.db --text=1 >signature-2n9.ckp
[00:00:00] Copying signature section
[00:00:00] Copying imprint section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] Generating candidates for 2n9
[00:00:00] numSlot=9 pure=0 numNode=2 interleave=504 numCandidate=424 numSignature=110(96%) numImprint=6327(95%) | skipDuplicate=0
[00:00:00] Sorting signatures
[00:00:00] Rebuilding imprints
[00:00:00] Imprints built. numImprint=6309(95%) | hash=1.095
[00:00:12] Writing signature-2n9.db
[00:00:12] Written signature-2n9.db, 854669088 bytes
[xyzzy@host v2.8.0]$ ./gensignature signature-2n9.db 3 signature-3n9.db --text=1 >signature-3n9.ckp
[00:00:00] Copying signature section
[00:00:00] Copying imprint section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] Generating candidates for 3n9
[00:00:02] numSlot=9 pure=0 numNode=3 interleave=504 numCandidate=81406 numSignature=5666(95%) numImprint=775137(95%) | skipDuplicate=5
[00:00:02] Sorting signatures
[00:00:02] Rebuilding imprints
[00:00:02] Imprints built. numImprint=773874(95%) | hash=1.102
[00:00:15] Writing signature-3n9.db
[00:00:15] Written signature-3n9.db, 932537216 bytes
[xyzzy@host v2.8.0]$ ./gensignature signature-3n9.db 4 signature-4n9.db --text=1 >signature-4n9.ckp
[00:00:00] Copying signature section
[00:00:00] Copying imprint section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] Generating candidates for 4n9
[00:05:45] numSlot=9 pure=0 numNode=4 interleave=504 numCandidate=29990974 numSignature=791647(95%) numImprint=181855572(95%) | skipDuplicate=2906
[00:05:45] Sorting signatures
[00:05:48] Rebuilding imprints
[00:06:40] Imprints built. numImprint=181679064(95%) | hash=1.145
[00:06:47] Writing signature-4n9.db
[00:06:54] Written signature-4n9.db, 19252524576 bytes
Section swap (2 minutes)
Single pass, no checkpointing.
[xyzzy@host v2.8.0]$ ./genswap signature-4n9.db swap-4n9.db
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] Generating swaps.
[00:01:47] numSwap=151(86%) | skipDuplicate=101518
[00:01:55] Writing swap-4n9.db
[00:02:01] Written swap-4n9.db, 19252534176 bytes
Section member (90 minutes)
Multi-pass upto 4n9. 5n9 is a beast and best fed to the grid.
NOTE: This section must have zero empty and unsafe members to be complete.
[xyzzy@host v2.8.0]$ ./genmember swap-4n9.db 1 member-1n9.db --text=1 >member-1n9.ckp
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] numImprint=181679064(100%) numMember=1(0%) numEmpty=791646 numUnsafe=0
[00:00:00] Generating candidates for 0n9
[00:00:00] numSlot=9 pure=0 numNode=0 numCandidate=0 numMember=3(0%) numPair=1(0%) numEmpty=791644 numUnsafe=0 | skipDuplicate=0 skipSize=0 skipUnsafe=0
[00:00:00] Generating candidates for 1n9
[00:00:00] numSlot=9 pure=0 numNode=1 numCandidate=6 numMember=9(0%) numPair=5(0%) numEmpty=791638 numUnsafe=0 | skipDuplicate=0 skipSize=0 skipUnsafe=0
[00:00:00] Sorting members
[00:00:00] Indexing members
[00:00:00] Indexed members. numMember=9 skipUnsafe=0
[00:00:00] WARNING: 791638 empty and 791638 unsafe signature groups
[00:00:00] {"numSlot":9,"pure":0,"interleave":504,"numNode":1,"numImprint":181679064,"numSignature":791647,"numMember":9,"numEmpty":791638,"numUnsafe":791638,"numPair":5}
[00:00:07] Writing member-1n9.db
[00:00:14] Written member-1n9.db, 19383506336 bytes
[xyzzy@host v2.8.0]$ ./genmember member-1n9.db 2 member-2n9.db --text=1 >member-2n9.ckp
[00:00:00] Copying pair section
[00:00:00] Copying member section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] numImprint=181679064(100%) numMember=9(0%) numEmpty=791638 numUnsafe=0
[00:00:00] Generating candidates for 2n9
[00:00:00] numSlot=9 pure=0 numNode=2 numCandidate=424 numMember=275(0%) numPair=13(0%) numEmpty=791537 numUnsafe=0 | skipDuplicate=0 skipSize=158 skipUnsafe=0
[00:00:00] Sorting members
[00:00:00] Indexing members
[00:00:00] Indexed members. numMember=275 skipUnsafe=0
[00:00:00] WARNING: 791537 empty and 791537 unsafe signature groups
[00:00:00] {"numSlot":9,"pure":0,"interleave":504,"numNode":2,"numImprint":181679064,"numSignature":791647,"numMember":275,"numEmpty":791537,"numUnsafe":791537,"numPair":13}
[00:00:07] Writing member-2n9.db
[00:01:43] Written member-2n9.db, 19383525536 bytes
[xyzzy@host v2.8.0]$ ./genmember member-2n9.db 3 member-3n9.db --text=1 >member-3n9.ckp
[00:00:00] Copying pair section
[00:00:00] Copying member section
[00:00:01] Rebuilding indices
[00:00:01] Indices built
[00:00:01] numImprint=181679064(100%) numMember=275(0%) numEmpty=791537 numUnsafe=0
[00:00:01] Generating candidates for 3n9
[00:00:04] numSlot=9 pure=0 numNode=3 numCandidate=81406 numMember=30079(0%) numPair=377(1%) numEmpty=785981 numUnsafe=0 | skipDuplicate=173 skipSize=51060 skipUnsafe=0
[00:00:04] Sorting members
[00:00:04] Indexing members
[00:00:04] Indexed members. numMember=30079 skipUnsafe=0
[00:00:04] WARNING: 785981 empty and 785981 unsafe signature groups
[00:00:04] {"numSlot":9,"pure":0,"interleave":504,"numNode":3,"numImprint":181679064,"numSignature":791647,"numMember":30079,"numEmpty":785981,"numUnsafe":785981,"numPair":377}
[00:00:15] Writing member-3n9.db
[00:01:35] Written member-3n9.db, 19385674336 bytes
[xyzzy@host v2.8.0]$ ./genmember member-3n9.db 4 member-4n9.db --text=1 >member-4n9.ckp
[00:00:00] Copying pair section
[00:00:00] Copying member section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] numImprint=181679064(100%) numMember=30079(0%) numEmpty=785981 numUnsafe=0
[00:00:00] Generating candidates for 4n9
[00:03:55] numSlot=9 pure=0 numNode=4 numCandidate=29990974 numMember=6183205(95%) numPair=48967(95%) numEmpty=0 numUnsafe=32 | skipDuplicate=57940 skipSize=22827233 skipUnsafe=119522
[00:03:55] Sorting members
[00:04:21] Indexing members
[00:04:26] Indexed members. numMember=6183205 skipUnsafe=0
[00:04:26] WARNING: 0 empty and 32 unsafe signature groups
[00:04:26] {"numSlot":9,"pure":0,"interleave":504,"numNode":4,"numImprint":181679064,"numSignature":791647,"numMember":6183205,"numEmpty":0,"numUnsafe":32,"numPair":48967}
[00:04:33] Writing member-4n9.db
[00:04:40] Written member-4n9.db, 19829088128 bytes
There are 32 incomplete signature groups so the scanning of 5n9 space is necessary.
Instead of dragnetting the entire space, search selectively for the missing members with a faster full associative index.
For that we need to create a specialised database.
First get a list of incomplete signature groups.
[xyzzy@host v2.8.0]$ ./genmember member-4n9.db 4 --no-generate --listunsafe >missing-4n9.lst
[xyzzy@host v2.8.0]$ wc missing-4n9.lst # should output 32
Then create a database with full associative lookups. This needs user supplied limits, each signature requires roughly worstcase 400000 imprints.
[xyzzy@host v2.8.0]$ ./gensignature transform.db 4 missing-4n9.db --load=missing-4n9.lst --no-generate --interleave=362880 --maximprint=150000000
Search 5n9 space, save the checkpoints and reapply them to the main database.
This can be done either single-cpu or with the grid.
Using single-core (50 minutes):
[xyzzy@host v2.8.0]$ ./genmember missing-4n9.db 5 --text=1 >missing-5n9.ckp
Using grid with 81 jobs/tasks:
[xyzzy@host v2.8.0]$ mkdir log-missing-5n9
[xyzzy@host v2.8.0]$ qsub -wd $PWD -o log-missing-5n9 -e log-missing-5n9 -b y -t 1-81 -q 4G.q -ckpt check ./genmember --task=sge missing-4n9.db 5 --text=1
[xyzzy@host v2.8.0]$ grep done log-missing-5n9/genmember.e* | wc # should output 81
[xyzzy@host v2.8.0]$ cat log-missing-5n9/genmember.o* >missing-5n9.ckp
If a grid is unavailable, and there is sufficient memory, then the option --task may give opportunity to run multi-core.
[xyzzy@host v2.8.0]$ ./genmember missing-4n9.db 5 --text=1 >>missing-5n9.ckp --timer=30 --task=1,4 &
[xyzzy@host v2.8.0]$ ./genmember missing-4n9.db 5 --text=1 >>missing-5n9.ckp --timer=30 --task=2,4 &
[xyzzy@host v2.8.0]$ ./genmember missing-4n9.db 5 --text=1 >>missing-5n9.ckp --timer=30 --task=3,4 &
[xyzzy@host v2.8.0]$ ./genmember missing-4n9.db 5 --text=1 >>missing-5n9.ckp --timer=30 --task=4,4 &
Then, finally, reapply the checkpoint data to the final step of this stage creation:
[xyzzy@host v2.8.0]$ ./genmember member-4n9.db 5 member-5n9.db --load=missing-5n9.ckp --no-generate --text=1 >member-5n9.ckp
[00:00:00] Copying pair section
[00:00:00] Copying member section
[00:00:00] Rebuilding indices
[00:00:01] Indices built
[00:00:01] numImprint=181679064(100%) numMember=6183205(17%) numEmpty=0 numUnsafe=32
[00:00:01] Reading members from file
[00:00:03] Read 124424 members. numSignature=791647(95%) numMember=6253958(18%) numPair=82653(4%) numEmpty=0 numUnsafe=0 | skipDuplicate=2647 skipSize=0 skipUnsafe=50970
[00:00:03] Sorting members
[00:00:17] Indexing members
[00:00:21] Indexed members. numMember=6253958 skipUnsafe=0
[00:00:21] {"numSlot":9,"pure":0,"interleave":504,"numNode":5,"numImprint":181679064,"numSignature":791647,"numMember":6253958,"numEmpty":0,"numUnsafe":0,"numPair":67925}
[00:00:32] Writing member-5n9.db
[00:02:10] Written member-5n9.db, 20459768640 bytes
Section complete with numEmpty=0 and numUnsafe=0.
Remove depreciated members (30 minutes)
Now all signature groups are complete can they be pruned.
Search and remove component members that do not contribute to uniqueness.
1n9 are root components, 2n9 are locked components, so start at 3n9.
[xyzzy@host v2.8.0]$ ./gendepreciate member-5n9.db 3 depr-3n9.db --text=1 >depr-3n9.ckp
[00:00:00] Copying signature section
[00:00:00] Rebuilding indices
[00:00:00] Indices built
[00:00:00] numMember=6253958(95%) numEmpty=0 numUnsafe=0
[00:00:01] numComponent=109080 numDepr=0 numLocked=303776
[00:00:10] numComponent=98109 numDepr=871782 numLocked=307154 | cntDepr=72 cntLock=36
[00:00:10] numComponent=98109 numDepr=871782 numLocked=307154
[00:00:21] Writing depr-3n9.db
[00:00:47] Written depr-3n9.db, 19878516928 bytes
[xyzzy@host v2.8.0]$ ./gendepreciate depr-3n9.db 4 depr-4n9.db --text=1 >depr-4n9.ckp
[00:00:00] Copying signature section
[00:00:00] Rebuilding indices
[00:00:01] Indices built
[00:00:01] numMember=5382176(95%) numEmpty=0 numUnsafe=0
[00:00:02] numComponent=95999 numDepr=0 numLocked=307154
[00:10:21] numComponent=73185 numDepr=1649868 numLocked=331359 | cntDepr=8433 cntLock=2224
[00:10:22] numComponent=73185 numDepr=1649868 numLocked=331359
[00:10:28] Writing depr-4n9.db
[00:10:35] Written depr-4n9.db, 19797364800 bytes
[xyzzy@host v2.8.0]$ ./gendepreciate depr-4n9.db 5 depr-5n9.db --text=1 >depr-5n9.ckp
[00:00:00] Copying signature section
[00:00:07] Rebuilding indices
[00:00:08] Indices built
[00:00:08] numMember=3732308(95%) numEmpty=0 numUnsafe=0
[00:00:08] numComponent=66856 numDepr=0 numLocked=331359
[00:00:08] 4( 2/s) 0.00886% eta=6:16:07 | numComponent=66852 numDepr=121 | cntDepr=4 cntLock=0 | refcnt=25 mid=86306 fab>ebcd!^!
[00:00:09] 7( 2/s) 0.01551% eta=6:16:05 | numComponent=66849 numDepr=199 | cntDepr=7 cntLock=0 | refcnt=23 mid=89084 fdabc!^ea>!
[00:00:09] 13( 4/s) 0.02880% eta=3:08:01 | numComponent=66843 numDepr=343 | cntDepr=13 cntLock=0 | refcnt=22 mid=89052 fdabc!^ae+!
[00:00:09] 21( 6/s) 0.04652% eta=2:05:19 | numComponent=66835 numDepr=527 | cntDepr=21 cntLock=0 | refcnt=21 mid=678938 babc!fade!^!
[00:00:09] 42( 10/s) 0.09305% eta=1:15:09 | numComponent=66814 numDepr=989 | cntDepr=42 cntLock=0 | refcnt=20 mid=523598 cdab^?fce>?
[00:00:09] 57( 12/s) 0.12628% eta=1:02:36 | numComponent=66799 numDepr=1304 | cntDepr=57 cntLock=0 | refcnt=19 mid=95139 fcab^d!ce+!
[00:00:09] 59( 8/s) 0.13071% eta=1:33:54 | numComponent=66797 numDepr=1344 | cntDepr=59 cntLock=0 | refcnt=18 mid=86622 fab>bcd^e!!
[00:00:09] 79( 12/s) 0.17502% eta=1:02:34 | numComponent=66777 numDepr=1724 | cntDepr=79 cntLock=0 | refcnt=17 mid=284960 fdabc!^ae+?
[00:00:09] 87( 11/s) 0.19274% eta=1:08:15 | numComponent=66769 numDepr=1868 | cntDepr=87 cntLock=0 | refcnt=16 mid=111743 abc!ade!e21!!
[00:00:09] 123( 17/s) 0.27250% eta=0:44:07 | numComponent=66733 numDepr=2480 | cntDepr=123 cntLock=0 | refcnt=15 mid=178010 fcdab^!ce&!
[00:00:10] 139( 13/s) 0.30794% eta=0:57:41 | numComponent=66721 numDepr=2672 | cntDepr=135 cntLock=4 | refcnt=14 mid=225704 cdab^!fce&!
[00:00:13] 215( 19/s) 0.47632% eta=0:39:24 | numComponent=66653 numDepr=3660 | cntDepr=203 cntLock=12 | refcnt=13 mid=90194 babc!ead>^!
[00:00:15] 293( 21/s) 0.64912% eta=0:35:35 | numComponent=66581 numDepr=4627 | cntDepr=275 cntLock=18 | refcnt=12 mid=85346 fab+eacd!^!
[00:00:17] 476( 29/s) 1.05454% eta=0:25:40 | numComponent=66406 numDepr=6851 | cntDepr=450 cntLock=26 | refcnt=11 mid=89657 ceabc!de!^3!
[00:00:20] 622( 31/s) 1.37800% eta=0:23:56 | numComponent=66268 numDepr=8359 | cntDepr=588 cntLock=34 | refcnt=10 mid=19232 ebdab>c1^!!
[00:00:23] 988( 52/s) 2.18884% eta=0:14:09 | numComponent=65910 numDepr=11753 | cntDepr=946 cntLock=42 | refcnt=9 mid=19222 ecab>c1d!^!
[00:00:27] 1288( 55/s) 2.85347% eta=0:13:17 | numComponent=65621 numDepr=14056 | cntDepr=1235 cntLock=53 | refcnt=8 mid=19199 edab+cd>^!
[00:00:33] 1903( 75/s) 4.21596% eta=0:09:36 | numComponent=65020 numDepr=18078 | cntDepr=1836 cntLock=67 | refcnt=7 mid=20059 bedabc!>^3!
[00:00:53] 2505( 31/s) 5.54965% eta=0:22:55 | numComponent=64478 numDepr=21143 | cntDepr=2378 cntLock=127 | refcnt=6 mid=19197 ebab+cd>^!
[00:01:22] 3822( 43/s) 8.46737% eta=0:16:00 | numComponent=63251 numDepr=25905 | cntDepr=3605 cntLock=217 | refcnt=5 mid=19236 ebdcab+^2!!
[00:01:51] 5014( 43/s) 11.10816% eta=0:15:33 | numComponent=62149 numDepr=29435 | cntDepr=4707 cntLock=307 | refcnt=4 mid=20069 ceabc!d>^3!
[00:02:58] 8196( 45/s) 18.15765% eta=0:13:40 | numComponent=59179 numDepr=36247 | cntDepr=7677 cntLock=519 | refcnt=3 mid=19209 ecab>cd>^!
[00:04:25] 11544( 28/s) 25.57490% eta=0:19:59 | numComponent=56098 numDepr=42189 | cntDepr=10758 cntLock=786 | refcnt=2 mid=19239 eadcab>^2!!
[00:08:49] 24605( 41/s) 54.51061% eta=0:08:20 | numComponent=43879 numDepr=60766 | cntDepr=22977 cntLock=1628 | refcnt=1 mid=18447 ab+eacd!+^
[00:17:33] numComponent=25102 numDepr=80933 numLocked=336892 | cntDepr=41754 cntLock=3384
[00:17:34] numComponent=25102 numDepr=80933 numLocked=336892
[00:17:45] Writing depr-5n9.db
[00:19:21] Written depr-5n9.db, 19643797344 bytes
Export and compress
Last step is to export the database (19.6GByte) into readable text/json (103MByte) and compress with xz (3.9MByte).
[xyzzy@host v2.8.0]$ ./genexport untangle-mixed.json depr-5n9.db
[xyzzy@host v2.8.0]$ xz untangle-mixed.json
untangle-pure.db from scratch (40 hours)
Steps similar to the mixed version, except adding the --pure option to all the program invocations.
This dataset is a tough cookie.
It takes 9 single-core hours to scan 5n9 space which finds all but 3 signature groups.
Scanning 6n9 space for the remaining 3 requires an additional 30 single-core hours.
In total some 33M members were found of which 381582 were selected as contributing.
./gentransform transform.db
#
./gensignature transform.db 1 signature-1n9.db --text=1 >signature-1n9.ckp --pure
./gensignature signature-1n9.db 2 signature-2n9.db --text=1 >signature-2n9.ckp --pure
./gensignature signature-2n9.db 3 signature-3n9.db --text=1 >signature-3n9.ckp --pure
./gensignature signature-3n9.db 4 signature-4n9.db --text=1 >signature-4n9.ckp --pure
#
./genswap signature-4n9.db swap-4n9.db --pure
#
./genmember swap-4n9.db 1 member-1n9.db --text=1 >member-1n9.ckp --pure
./genmember member-1n9.db 2 member-2n9.db --text=1 >member-2n9.ckp --pure
./genmember member-2n9.db 3 member-3n9.db --text=1 >member-3n9.ckp --pure
./genmember member-3n9.db 4 member-4n9.db --text=1 >member-4n9.ckp --pure
#
mkdir log-member-5n9
qsub -wd $PWD -o log-member-5n9 -e log-member-5n9 -b y -t 1-81 -q 4G.q -ckpt check ./genmember --task=sge member-4n9.db 5 --text=1 --pure
# wait-till-complete
grep done log-member-5n9/genmember.e* | wc
cat log-member-5n9/genmember.o* >member-5n9.ckp
./genmember member-4n9.db 5 member-5n9.db --load=member-5n9.ckp --no-generate --pure
#
./gensignature member-5n9.db 4 --no-generate --listunsafe >missing.lst
wc missing.lst
./gensignature transform.db 5 missing-5n9.db --load=missing.lst --no-generate --interleave=362880 --maximprint=12800000 --pure --maxsignature=90
#
mkdir log-missing-6n9
qsub -wd $PWD -o log-missing-6n9 -e log-missing-6n9 -b y -t 1-2187 -q 4G.q -ckpt check ./genmember --task=sge missing-5n9.db 6 --text=1 --pure
# wait-till-complete
grep done log-missing-6n9/genmember.e* | wc
cat log-missing-6n9/genmember.o* >member-6n9.ckp
./genmember member-5n9.db 6 member-6n9.db --load=member-6n9.ckp --no-generate --pure
Requirements
- Working
untangleenvironment. https://github.com/RockingShip/untangle - 32G memory minimum.
- Suggested: 64G with 8-core CPU.
- Suggested: fast DDR using four banks
- 512G SSD minimum.
- Sun Grid Engine, when building from scratch.
- Star Cluster, for running SGE within Amazon S3.
Versioning
We use SemVer for versioning. For the versions available, see the tags on this repository.
License
This project is licensed under the GNU AFFERO General Public License v3 - see the LICENSE.txt file for details